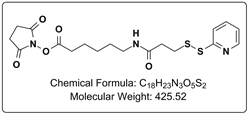
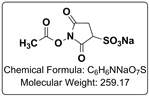
Crosslinking with DTSSP, separation with SDS-PAGE, digestion and subsequent MS analysis. DTSSP makes mass spectrometry analysis less complicated.Chemically crosslinking proteins, of a cell lysate, on the surface or inside of cells, with a crosslinker, such as BS3, are common ways to perform protein crosslinking reactions. After completion of the crosslinking reaction (and cell lysis if necessary), often the crosslinked protein complexes are identified through 2D-SDS-PAGE, in gel trypsin digestion, followed by mass spectrometry, such as MALDI-MS. This approach, can give direct evidence for specific protein-protein interactions, as it can be safely inferred that the two proteins would have to be interacting with one another in order to be crosslinked with a small molecule protein crosslinker. This approach can at times be cumbersome, and the mass spectrometry data generated can be overwhelming. One way to simplify the data analysis would be to use a cleavable crosslinker like CovaChem’s DTSSP. After the proteins have been crosslinked with DTSSP, the lysed mixture could be immunoprecipited for the protein of interest, the precipitate washed, then reduced with 10-50 mM DTT (CovaChem 11302-10×5), separated with SDS-PAGE, digested in-gel with trypsin, followed by MALDI-MS analysis. Using this approach with DTSSP, and cleaving the crosslink between the two proteins allows for easier identification of interacting proteins, as the proteins can be isolated from one another prior to digestion, and subsequent mass spectrometry. Sometimes a little simplification of the data can make all the difference. Best of luck in your research. |